Monday, July 1, 2024
Detritus / Wee Dude / Fairypoo Pumpkinpie / Little Princeling. 2009-2024
Tuesday, January 31, 2023
Farewell Mrs Spat
Tuesday, November 2, 2021
Cycle of cells
Here is a September 2020 retraction from Cell Cycle. The editors sound defensive and aggrieved, for reasons we can only guess in the absence of most of the story.
We, the Editor[s] and Publisher of Cell Cycle, have retracted the following article:
NEAT1 regulates Th2 cell development by targeting STAT6 for degradation
Shuman Huang, Dong Dong, Yaqian Zhang, Zhuo Chen, Jing Geng & Yulin Zhao
Pages 312 - 319 |https://doi.org/10.1080/15384101.2018.1562285
The above article has been retracted as a result of concerns regarding errors in the data by request of the authors.
We note that we received, peer-reviewed, accepted, and published the article in good faith based on the purported veracity of these representations and warranties. We have been informed in our decision-making by our policy on publishing ethics and integrity and the COPE guidelines on retractions.
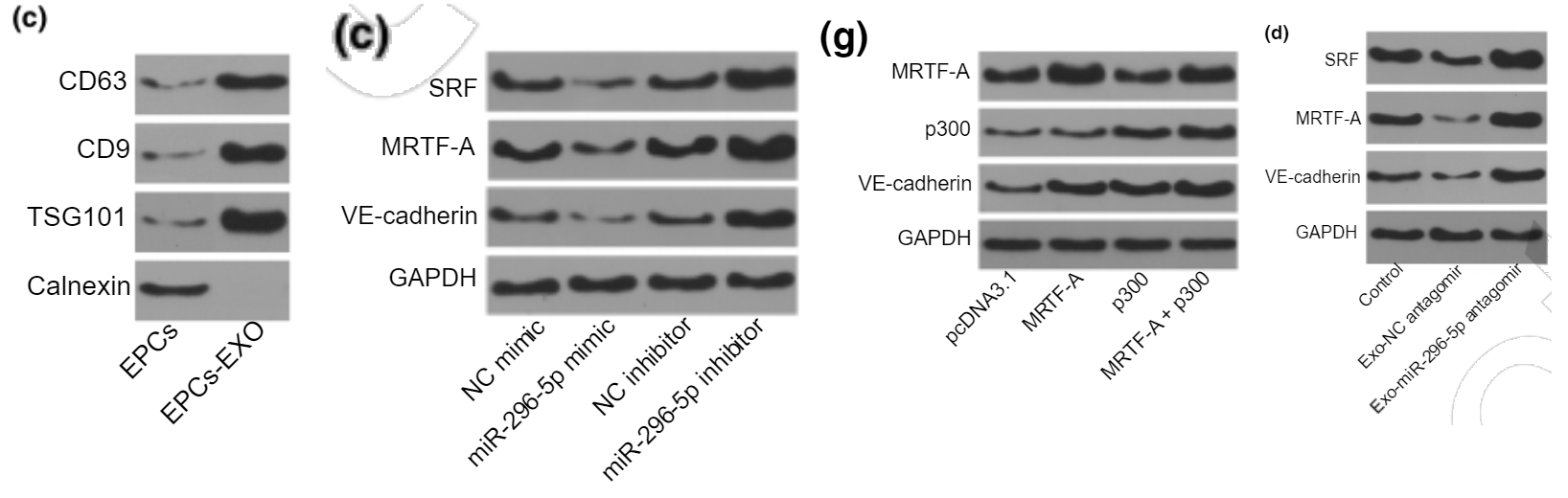
And a 2020 Corrigendum, in which the journal's editors drew a discreet veil over two Figures afflicted with PCD (Photoshop-Compulsion Disorder) and allowed the authors to replace them with better visual compositions. The conclusions were not affected.
It has been noted by the authors that the Figures 2 and 4 were published with errors. These errors have now been corrected as shown below. This correction has not changed the description, interpretation, or the original conclusions of the article. The authors apologize for any inconvenience caused.
This next retraction, at first glance unrelated to the first, comes from Cancer Cell International. The authors accepted that the experiments they reported, as well as the bio-informatic analyses of existing databases, were the uncredited work of an outside laboratory, which refuses to provide raw data for scrutiny and validation. Though authors were at pains to stress that they did collect some basic stats at their own institutions.
The Editors-in-Chief have retracted this article [1] at the request of the authors because concerns have been raised regarding validity of data.
The authors informed the journal that the bioinformatics analysis and all laboratory contents had been performed by an external company. The outsourced laboratory content includes cell culture, FISH, dual luciferase reporter gene assay, RNA pull down assay, RNA immunoprecipitation (RNA IP) assay, cell grouping and transfection, RT qPCR, western blot analysis, chemosensitivity assay and flow cytometry. This was not made clear in this article. The company has not responded to requests from authors to share raw data. Consequently, the data cannot be verified.
The authors stated that they completed the rest of the work, including clinical patient samples immunohistochemistry analysis, statistical analysis and discussion.
All authors agree to this retraction.
All this means that the stars are aligned for another scholarly inquiry into the Chinese papermill industry (an inexhausible topic, for me at least). I continue to be guided by the principle that a journal becomes a useful research tool when papermills take it over: a controlled environment in which their products can be studied in splendid isolation, without the complicating factor of genuine scholarship, revealing the shared, recurring elements and defining characteristics of specific studios. We will encounter more than one papermill, and more than one journal. There will be data-market speculation, and a short test afterwards to see who was paying attention. Here is a foretaste to motivate readers to continue:
The invasion-assay and colony-assay images from Dai et al (2019) are a kaleidoscope of unexpected similarities...
...but they should not distract our attention from the serried ranks of tumors, which overlap with those of Han et al (2019). Garbage lists of primers also overlap.
Figs 1c and 1d from Li et al (2020) are possibly the work of an adversarial generative neural net that someone trained on tissue slides.
From the same source, Figs 2b ("BEFORE") and 4c ("AFTER") show what happens to a rodent brain after you damage it with ischaemia / reperfusion, then cut it up and try to transmit the slices via a glitching fax.
I'm not sure whether Fig 3b from Zeng et al (2019) is a hommage to Fontana's Concetto Spaziale perforated canvases, but the colored annotations turn it into that style of sharp-shooting where you blaze away at the side of a barn and then draw a bullseye around each bullet-hole. You might see this as a metaphor for the methodology of cell biology but I couldn't possibly comment. At right, Fig 5a was probably fun to create, but it is for entertainment purposes only.
* * * * * * * * * * * * * * * * * * * * * *
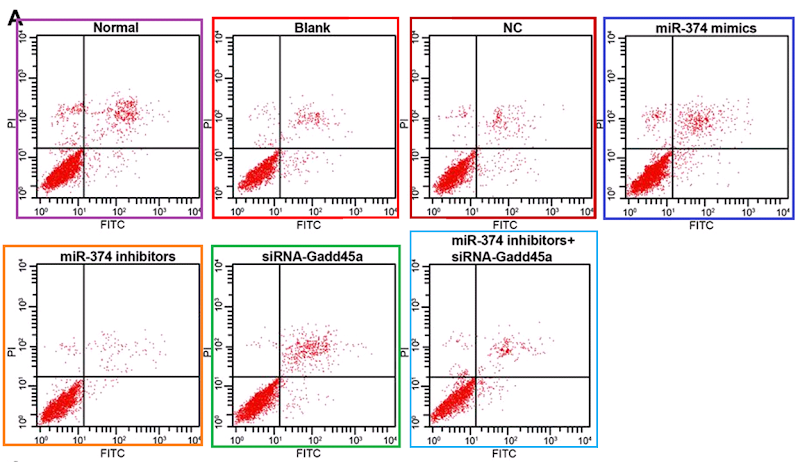
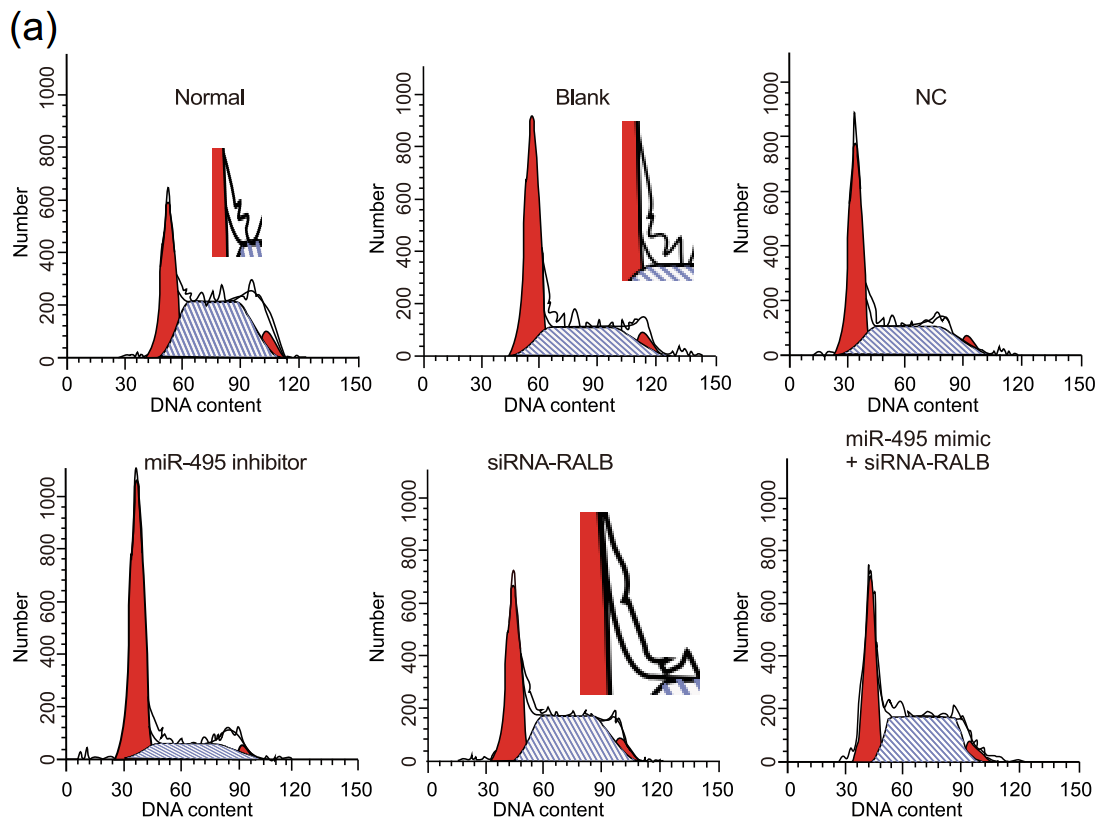
Anyway... these are flow-cytometry cell-cycle analyses. Like a Luc Besson movie, each panel consists of five elements:
- 1. An empirical distribution of DNA content per cell (as revealed though fluorescent markers), for a population of usually culture-grown cells. This is the wiggly zigzag line.
 2, 3, 4. Theoretical distributions of DNA content that one would expect from percentages of the population that were in the G0/G1, S or G2/M stages of the endless cycle of
2, 3, 4. Theoretical distributions of DNA content that one would expect from percentages of the population that were in the G0/G1, S or G2/M stages of the endless cycle of violencemitosis. These are the two crimson peaks bracketing a cross-hatched mesa to form a Feininger painting of a church.- 5. The smoother solid line hewing close to the zigzags is the sum of 2, 3 and 4; by adjusting their percentage parameters, it has been matched as closely as possible to 1.
These diagrams are relevant to the interests of oncology. Any treatment or drug that arrests the cycle (e.g. by alerting the DNA-integrity checkpoints at the transitions G1-to-S or G2-to-M) is a good thing from the perspective of a tumor's unwilling host, and its efficacy can be quantified by the accumulation of checkpointed cells in these plots (and their percentages).
Clearly, none of the curves 1 to 5 should overhang or stick out sideways like something left out on the sun or boiling over on the stove. It is also obvious that 5 is always as high as or higher than each of the components that it's the sum of... if it is hidden behind any of the humps, something has gone awry. Where the central mesa intersects either of the crimson peaks, their sum 5 is twice the height of that intersection.
And one more corollary: unless there are other parameters to fit, the shape of a given hump should be constant across all of the panels, only varying in height as it's multiplied by the percentage of cell population, fitted for each panel.
Cell Cycle is also a journal, published since mid-2014 by Taylor & Francis (i.e. Informa), with a hybrid model: papers are pay-to-read by default, unless authors paid to have them unlocked as free-to-read (all papers older than July 2020 seem to be free-to-read; I don't know if that reflects a change in policy, or the paywall is more of a one-year embargo).
Its founding Editor-in-Chief was Mikhail Blagosklonny, impresario of oncology literature and a serial founder of journals - he still occupies an honorary post on the Editorial Board despite stepping back from control. Mischa also gave us Aging and Oncotarget. He deserves credit for his early recognition of the academic-publishing opportunities created by (a) the shift away from subscription models to the open-access paradigm; and (b) the emergence of new areas for oncological endeavour, like exosomes and noncoding RNAs. (b) triggered a research-funding goldrush, while (a) motivated researchers to stake out their claims over regions of these ecologies of inter- and intra-cellular RNA signaling.
A 2008 'interview' focused on those aspects of science publishing that matter most: a high citation rate, and an abbreviated review cycle so that authors could win the race to announce breakthroughs. Blagosklonny ascribed Cell Cycle's success on both counts to his policy of prioritising a high-powered Editorial Board, to judge what is newsworthy, over peer reviewers with their focus on mere correctness.
How would you account for the high citation rate of Cell Cycle?
... Editors should not rubber-stamp reviewer’s decisions (otherwise, sometimes the best and most original papers will be rejected). Actually, reviewers should not decide at all whether a paper should be published or not. Reviewers provide criticism and reveal mistakes but should not decide a paper’s fate. .... For the most competitive papers, an ultra-rapid review (by members of the Editorial Board) is necessary to publish them a few days after submission.
His aspirations for the journal's citation indices have been fulfilled (with assistance from citational shenanigans, some have alleged). I hope this compensates for the extent to which Cell Cycle has become a vehicle for the papermill industry. Recent issues include (A) eight products from the celebrated 'Tadpole papermill'; (B) 15 from a "Directly Targeting" mill; and (C) at least 15 attributable to "Contractor" Redux, though there are undoubtedly more. Spreadsheets dedicated to those mills overlap with one I set up for present purposes, as is the custom of my people.
I concentrated on a two-year window, 2019-2020. Prior to this period of exuberance, the journal's Tables of Content are shorter, and have gone relatively untouched by the pseudonymous Pubpeer scrutineers (this might reflect a selection bias, or it could be that earlier papers were faked but in styles that are less obvious because less familiar). The editors were so delighted to receive an increased flux of submissions that they didn't wonder about the source. Hat-tip to Pubpeer contributor 'Indigofera Tanganyikensis', who was deconstructing Cell Cycle papers and questioning its editorial standards long before anyone else piled on. However, Indigofera's conclusion that everything in the journal is fictional is a slight exaggeration.
Unfortunately, this is apparently common practice for the publisher Taylor & Francis. They share no responsibility for the content of their publications. They know that Cell cycle is totally infiltrated by the fabrications from the different papermills from China, but there are apparently no initiative to stop this terrible mass disinformation.
(B) and (C) need their own subsections. For (A) a few examples will suffice. Wang et al (2020) and Zheng et al (2020) capture this mill when its distinctive 'zany sardine' WBs had evolved into avant-garde door-handles, designed perhaps by Philippe Starck.


Ren et al (2020) and Zhang et al (2020) have developed less from the original style.
Figs S1 to S3 from Ren, Li & Wang (2019) exemplify the wonderfully rococo but rigidly formalised "flaw cytometry" images created by this mill.
* * * * * * * * * * * * * * * * * * * *
I named this mill after the rigid title-generating template used in its early products, where the keystone of the title sentence was "by targeting X" or "by directly targeting Y". The template later accepted more diversity. Those early examples used Spandidos journals as hosts, but the mill later migrated to OncoTargets & Therapy and Cancer Management & Research from Dove Press, and then to Cell Cycle. Aging receives their patronage as well. Dove Press is itself another branch of the Taylor-&-Francis Borg (since 2017), but its editors are less averse to wielding the Hammer of Retraction.
This mill is easily characterised:
(a) Genuine Transwell invasion-assay images, from a small number of experiments, so the images sometimes overlap between papers. More overlaps await discovery by someone equipped with ImageTwin to take over the boring part of multiple comparisons.
(b) Western Blots resembling strings of sausages, which makes sense given the kinship of sausage factories and papermills.
 (c) ID parades of xenograft tumors excised from nude mice, though they are yellow and bloodless, and could be moulded lumps of Fimo or plasticine for all I know. The repeat appearances of peripatetic rulers, in studies from unrelated research teams at distant institutions, confirm that photographs were taken at some single laboratory (not necessarily linked to the papermill). Sometimes the tumors themselves recur between papers.
(c) ID parades of xenograft tumors excised from nude mice, though they are yellow and bloodless, and could be moulded lumps of Fimo or plasticine for all I know. The repeat appearances of peripatetic rulers, in studies from unrelated research teams at distant institutions, confirm that photographs were taken at some single laboratory (not necessarily linked to the papermill). Sometimes the tumors themselves recur between papers.
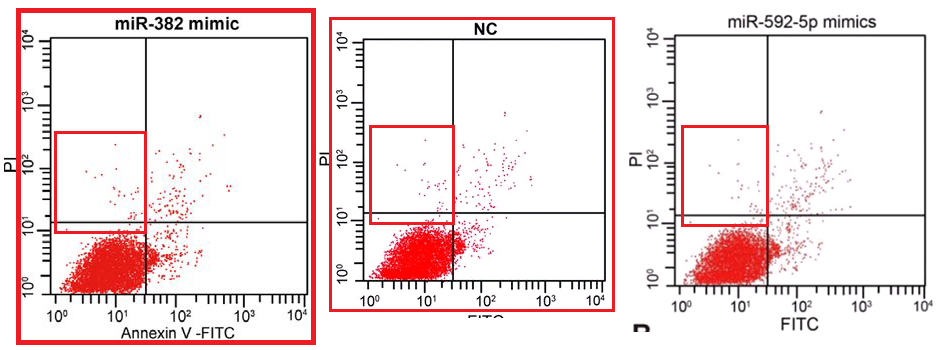
(d) Flow-cytometry scatterplots, in which specific combinations of PI and Annexin signals indicate which cells in a population are self-destructing (apoptotic)... again, the proportion of apoptosis in response to a drug treatment is of interest to cancer patients.
 These last are hard to fake convincingly (though 'plausibility' is a low priority in the literature). The problem is their stochastic nature. Humans are really crap at drawing stochastic distributions, even when equipped with a Photoshop stipple brush, and we cannot help spacing out the dots evenly, with results that at worst resemble a partridge. In the real world, though the coordinates of cells along the two fluorescence axes are guided by the population's overall distribution, each cell is oblivious to the locations of other cells; they neither seek nor avoid one another's proximity in the scatterplot.
These last are hard to fake convincingly (though 'plausibility' is a low priority in the literature). The problem is their stochastic nature. Humans are really crap at drawing stochastic distributions, even when equipped with a Photoshop stipple brush, and we cannot help spacing out the dots evenly, with results that at worst resemble a partridge. In the real world, though the coordinates of cells along the two fluorescence axes are guided by the population's overall distribution, each cell is oblivious to the locations of other cells; they neither seek nor avoid one another's proximity in the scatterplot.
The Targeting studio was fortunate enough to acquire a suite of bona-fide data-sets from multiple experiments with multiple replications of each experiment, allowing them to illustrate their productions by grabbing plots at random. They range from harps to banner-waving beavers. It's a large suite, but the studio was productive enough that panels did get re-used (sometimes plotted with filters or altered gate settings to look slightly different).
Then they acquired a second experimental corpus, sampling another niche of "shape space" with plots that bring musical bass-clefs to mind, or the sigmata of integration, or amphoras. Amphorices.
I infer that channels exist whereby lab technicians (and students, and perhaps the PIs) can sell their laboratories' image archives to mills, for which they are valuable visual material - lending verisimilitude to otherwise-bare assertions about numerical results. Any information about this hypothetical Goblin Market would be welcome.
 One author showed up at Pubpeer as rightful owner of a suite of Porcupine / Thidwick-themed FC plots that had been sampled and resampled to illustrate the oeuvre of the 'Traveling Tumor' papermill. It seemed that students had absconded with the experimental database, explaining its escape into the wild. No-one else claimed to be the source so I am happy to accept that researcher's priority.
One author showed up at Pubpeer as rightful owner of a suite of Porcupine / Thidwick-themed FC plots that had been sampled and resampled to illustrate the oeuvre of the 'Traveling Tumor' papermill. It seemed that students had absconded with the experimental database, explaining its escape into the wild. No-one else claimed to be the source so I am happy to accept that researcher's priority.
Dear Hoya Camphorifolia
Thank you for your comments to our article, regarding these flow cytometry images. The experiments of this study was stared in 2014 and finished in 2018. After carefully checking, we noticed that some of our data, including these flow cytometry images, might have been divulged to other researchers by our graduated students, and had been published. We had contacted the authors in the articles as you mentioned. We confirmed that these articles had been or are being retracted by the corresponding authors. I apologize for any confusion this may have caused to you and other readers.
Sincerely yours Songbai Yang
(C) Contractor
This mill has featured on Forbetterscience twice already. It provided clinicians with bespoke solutions to their publication / promotion problems, with manuscripts tailored to the nominal authors' specialties, perhaps building on or even completing some research of their own. In a recent retraction purge, 76 of its productions were de-published from the Journal of Cellular Biochemistry, and 32 from the J. of Cellular Physiology. Which is to say, the editors and the management at Wiley accepted the arguments marshalled in Pubpeer threads that all these papers displayed the hallmarks of the Contractor style. In some retractions, the criticism had not progressed as far as a Pubpeer thread but the papers were included in my spreadsheet, suggesting that the editors consulted the spreadsheet in the course of their inquiry.
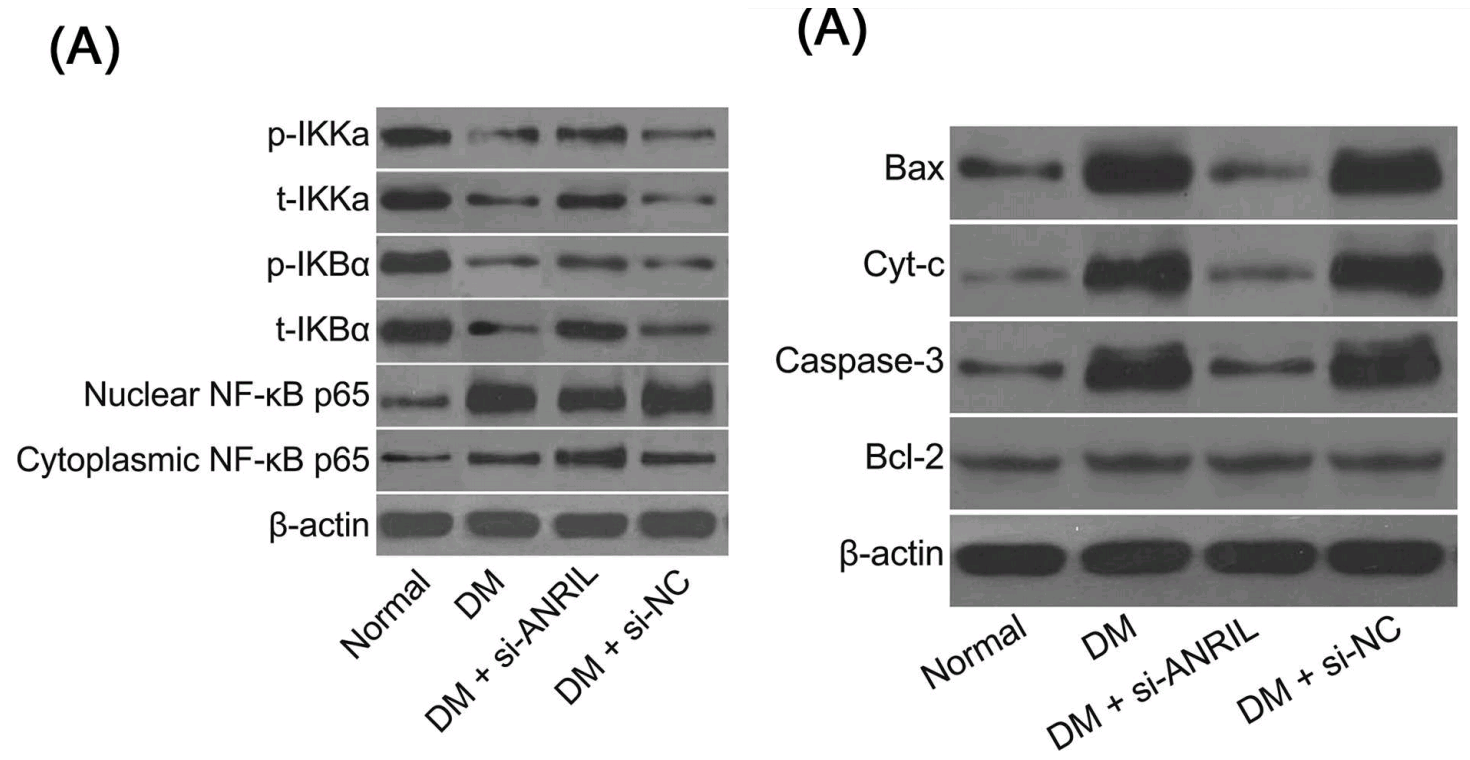
 The defining hallmarks used to be a versatile troupe of Spiny-Norman / Champagne-bottle FC apoptosis diagrams, that might once have emerged from actual experiments before endless recycling and modification erased all trace of their origins; and an alphabet of Western-Blotty protein blobs, cut up and reassembled ransom-note fashion into a kaleidoscopic supply of WB bands that were at the same time inexhaustible yet always familiar.
The defining hallmarks used to be a versatile troupe of Spiny-Norman / Champagne-bottle FC apoptosis diagrams, that might once have emerged from actual experiments before endless recycling and modification erased all trace of their origins; and an alphabet of Western-Blotty protein blobs, cut up and reassembled ransom-note fashion into a kaleidoscopic supply of WB bands that were at the same time inexhaustible yet always familiar.
Slices of brain or heart tissue, brightly coloured with the classic histology stains or with the antibodies of immunochemistry, sometimes repeated across productions as well. Let us draw a sympathetic veil over the occasions when slices of rodent cerebrum illustrated experiments on myocardial infarction.
 Also, Flaw-Cytometry cell-cycle histograms, which should be the easiest of all to fake convincingly. It is evident, though, that many of these images were created by people with photoshop skills but no grasp of the principles that undergird FC and govern how the elements fit together. I choose to believe that these were the work of interns: new recruits to the studio, gaining on-the-job training. If they had a cell-biology background they'd be in another office, assembling the text, but instead they were handed a stack of figures from publications that fooled the reviewers, and told to 'make more like this'. I can sympathise, having no background in the Dark Arts either: all this pontification is based on what I intuit from looking at examples, plus dim memories of reading Herzenberg's 1976 introduction to the exciting new technology of FACS.
Also, Flaw-Cytometry cell-cycle histograms, which should be the easiest of all to fake convincingly. It is evident, though, that many of these images were created by people with photoshop skills but no grasp of the principles that undergird FC and govern how the elements fit together. I choose to believe that these were the work of interns: new recruits to the studio, gaining on-the-job training. If they had a cell-biology background they'd be in another office, assembling the text, but instead they were handed a stack of figures from publications that fooled the reviewers, and told to 'make more like this'. I can sympathise, having no background in the Dark Arts either: all this pontification is based on what I intuit from looking at examples, plus dim memories of reading Herzenberg's 1976 introduction to the exciting new technology of FACS.
Later the studio phased out its limited WB gamut, upgrading to new alphabets (or, it may be, acquiring fake-blot algorithms in which simple brushstrokes are variously filtered, degraded, and overlaid onto noise-texture backgrounds, with results that are repetitive but not provably repetitions). We meet grassgrub larvae, and Christmas crackers, and another version of the sausage-link style... bands in each of these styles maintain their morphology, not progressively developing as the putative protein advances along the putative gel. There is room for the creation of a more rigorous taxonomy (but not the inclination). Editors are beginning to see the difference between Western Blotting and sausage manufacture, so that particular style is losing popularity in turn.
These new illustrations accompanied the mill into the pages of Cell Cycle. The millers also lifted their FC game to make their apoptosis scatterplots less mannered and recognisable. Anyone who's read Mallory Ortberg's contributions at The Toast will imagine the studio environment in the voices of Two Monks Inventing Things:
- what should a cell-cycle plot look like
- a long stripy mound with a red spike at each end. then you draw a wriggly line along the top to keep the birds off
- this one came out like a church on fire
- yes that's good
- how about an apoptosis scatterplot
 like a bursting puffball. or a sneezing sphinx, but its mother was a dust-bunny so it's made of little dots.
like a bursting puffball. or a sneezing sphinx, but its mother was a dust-bunny so it's made of little dots.- all those dots take a long time to draw
- it's faster if you use the stipple tool
- no-one even told me what apoptosis is
- it's the name of an Egyptian pharaoh
But flow cytometry remains the Achilles heel of the Contractor mill, with faults that come to light with the passage of time, in line with the old adage that "Time wounds all heels".
The mill added bioinformational analysis to the repertoire: number-crunching available data to form networks or Venn diagrams, which emanate geometrical clarity and imply that the inchoate information-overload of protein / ncRNA interactions conceals a simplicity that holds sway above the flux. This bioinformational flim-flam may well be valid and unimpeachable, in a GIGO way, but still 100% meaningless.
Crucially, these extensions to the repertoire are not limited to Cell Cycle. We find the same interaction tools with the same display-option settings, the same WB styles and heuristics for hand-drawn FC scatterplots, throughout the Springer / Nature stable of journals, and the Biomedcentral stable "evolving portfolio of some 300 peer-reviewed journals" (now assimilated into Springer / Nature). There are hundreds more of these productions, and probably thousands. This is the unspoken corollary to the retraction of Gao et al (2020): all the other papers with this general pattern is also the work of the same outside company. If that company were to suddenly own up to malfeasance, great swathes of attested fact in the fields of miRNA and Exosomes would just evaporate.
The authors informed the journal that the bioinformatics analysis and all laboratory contents had been performed by an external company. The outsourced laboratory content includes cell culture, FISH, dual luciferase reporter gene assay, RNA pull down assay, RNA immunoprecipitation (RNA IP) assay, cell grouping and transfection, RT qPCR, western blot analysis, chemosensitivity assay and flow cytometry.
All that's a lot to process. But there's more! I have saved the new stuff until last, to reward readers who persevered with this post. Whole classes of bodgy Cell Cycle papers remain. That single Retraction looks increasingly lonely as the number of Pubpeer threads addressing Cell Cycle papers passes 100 (while authors decline, or didn't receive, the invitations to participate in discussion). Perhaps the Editors feel they are absolved of responsibility by the Author Contributions paragraph of each fabrication, which nominate one author to stand as Guarantor of Data Integrity.
Illustrators have been spurred to greater ingenuity, with image manipulation for its own sake (when simply stealing from elsewhere would have been enough), as if they had signed onto a Photoshop Challenge of which we are the beneficiaries. Hand-crafted Transmission Electron Microscopy abounds.
 Yi et al (2019) came up with a table of phony numbers, and another artisanal TEM. But their Figure 1c is what catches the eye. After so much time was invested in these compositions, cutting and pasting, smoothing and sponging, it would be the grossest ingratitude to just skim over them without any attempt at deconstruction.
Yi et al (2019) came up with a table of phony numbers, and another artisanal TEM. But their Figure 1c is what catches the eye. After so much time was invested in these compositions, cutting and pasting, smoothing and sponging, it would be the grossest ingratitude to just skim over them without any attempt at deconstruction.
Moving right along, consider the following oddly-worded Acknowledgement:
We would like to acknowledge the reviewers for their helpful comments on this pap
The pap-related choice of words seems unlikely to occur to many authors independently; yet it features in seven Cell Cycle papers, as another symptom of a papermill provenance. These include Rong et al (2020):
 There isn't really enough room for the images from Tian et al (2020), but they are too good to leave out. They begin by mapping out possible landing sites on satellite photographs of Mars; then things get weird.
There isn't really enough room for the images from Tian et al (2020), but they are too good to leave out. They begin by mapping out possible landing sites on satellite photographs of Mars; then things get weird.
I am drawn towards the fake rat-brain slices that abound in this oeuvre, on account of the delightful color palette involved in imitating hematoxylin-eosin staining. Fig 2a of Liu et al (2020) offer hidden symmetries as well!
Li et al (2020). One can only imagine the implausibility of these images before the reviewers provided helpful suggestions for their improvement.
The unabbreviated version would be unexceptional, in isolation, but it accompanies other symptoms in 33 papers.
We would like to acknowledge the reviewers for their helpful comments on this paper
These include Liu et al (2020), whose Figure 3 is teeming with so much Bikbait that I hardly know where to begin.
Li et al (2020) offer Fig 5e, "the results of electron microscopy observation of the [femoral heads] of rats in each group".
Please admire the fearful symmetries within Figure 1b of Li et al.
 These examples are an invitation to enter a whole new rabbit-warren of fulsome Acknowledgements. Many are courteous words that authors might easily borrow from elsewhere, not trusting their imperfect English to properly state their gratitude to colleagues and reviewers...
These examples are an invitation to enter a whole new rabbit-warren of fulsome Acknowledgements. Many are courteous words that authors might easily borrow from elsewhere, not trusting their imperfect English to properly state their gratitude to colleagues and reviewers...
The authors would like to acknowledge the helpful comments on this paper received from the reviewers.
We send our sincere gratitude to the reviewers for their kind comments
We thank the reviewers for critical comments
We would like show sincere appreciation to the reviewers for critical comments on this article (x 2)
We would like to acknowledge the helpful comments on this paper received from our reviewers (x 2)
We would like to acknowledge the reviewers for their helpful comments on this study
We would like to express our gratitude for the helpful comments received from our reviewers.
We would like to give our sincere appreciation to the reviewers for their helpful comments on this article
We would like to acknowledge the reviewers for their helpful comments on this paper
... But they appear in other Contractor productions in journals from elsewhere in the oeuvre, even leading to journals that hitherto had gone unexamined. One style of WB fabrication was previously unfamiliar, with bands like a display-case in a prosthetic-eyebrow salon... sometimes in combination with apoptosis scatterplots modeled after comets, or fried eggs (with a sharp edge around the 'white' and another between the 'white' and the 'yolk').
Figs 1b, 7a of Yang et al (2021)
Figs 5f,h of Wang et al (2020)
And what is one to make of Acknowledgements that address the authors, in third-person?
We are appreciating to every author for their assistance with the experiments and constructive comments on the manuscript (x 2)
We acknowledge and appreciate our colleagues for their valuable efforts and comments on this paper (x 3)
We would like to thank our researchers for their hard work and reviewers for their valuable advice.
Why not name those anonymous colleagues whose contributions were substantial enough to earn a credit, but not enough for co-authorship? Is this a sly wink from the scriptorium monks, a joke about their invisibility?
 One last Contractor production, Fig 2a in Luo et al (2019), appeared in Cancer Cell International rather than Cell Cycle - but I finish with it anyway as a pinnacle of their art.
One last Contractor production, Fig 2a in Luo et al (2019), appeared in Cancer Cell International rather than Cell Cycle - but I finish with it anyway as a pinnacle of their art.
UPDATE: The authors of 1% of the Cell Cycle papers in question have responded to the invitation to join the discussion. The explanation for the symmetries and clone-stamp irreality of their images was not entirely satisfying.
Thank you for your attention to our manuscript and helpful suggestions. I’m writing to you in response to the inquiries you posted with regard to the study “Overexpressed microRNA-494 represses RIPK1 to attenuate hippocampal neuron injury in epilepsy rats by inactivating the NF-κB signaling pathway”. Firstly, after carefully rechecking all the images again, we have figured out that this error was caused due to my negligence during the figure assembly as there were too many very similar figures. When we cropped the original images to collectively present the representative pictures, we accidentally inserted the wrong images into the different groups, leading to some overlap and similar pictures between and within groups. Besides, the problems mentioned in Figure 2 might also be resulted from a stutter of the imaging system.
Luo et al (2019) have belated doubts about their Figure 2a and have decided to withdraw the paper.
Thank you for your attention to our research and the questioning of some of the experimental pictures. We attach great importance to this news and have carried out the verification of the original experimental data as soon as possible. There are indeed some detailed problems. Due to a long period of time, the original records of the electron microscope room are no longer available. In order to further clarify the research conclusions, we plan to repeat the experiment. However, considering the repetition of the electron microscope experiment, it takes a long time to avoid misleading and troublesome to other researchers. All authors agree that they have decided to contact the journal editorial department to apply for withdrawal of the manuscript. In future research, we will pay attention to details to ensure scientific research.
* Three groups are noted for their Dunning-Kruger confidence when they saunter blithely into some other magisterium to give those experts the benefit of their superior insight, unimpeded by the blind-spots acquired through training or experience: physicists, economists and software coders.