Additions by Tiger. This post was earlier cross-posted at Leonid Schneider's site, hence the nonfrivolity and Explaining Voice. The version there is improved by Leonid's editing and explanation of the back-story, and readers' comments.
Feel free to admire these panels. The angular silhouettes are not cranes in flight or fish schooling among bubbles, but vat-cultured carcinoma cells from a lineage prone to metastases; they have been lured through pores in a microfilter and then stained, to quantify how different conditions and deterrents affected their invasive tendencies. Sadly in light of the positive results of this 'migration assay', it did not exist outside Photoshop, and in comments at PubPeer, 'Bacillus Mannanilyticus' has helpfully circled some of the clone-tool-multiplied cells.
The authors took the criticism to heart and recently retracted the paper --
This article has been withdrawn at the request of the authors. It contained several inappropriately processed and incorrect figures, Figs. 3C, 3D, 4B, 4C, 4D, 5B, 5D, 6B, 7B, 7C, and 8B. On behalf of all authors, the corresponding author has taken full responsibility and apologizes to the readers of Molecular & Cellular Proteomics for submitting and publishing the erroneous article and any inconvenience caused.-- though without crediting PubPeer or B. Mannanilyticus... perhaps they learned of the cloning accidents through other channels.
An earlier retraction from the same research group hinged on overlapping microphotographs. Let us say that the number of stained tissue slices (and thus the number of distinct treatments) represented in Figures 5 and 7 was smaller than the number of panels in those subdivided diagrams. Like roofing tiles, or Ordnance Survey maps, the panels share many edges in common.
This article has been retracted at the request of the authors. It contained several inappropriately processed and incorrect Figures. On behalf of all authors, the corresponding author has taken full responsibility and apologizes to the readers of BBA Molecular Basis of Disease for submitting and publishing the erroneous article and any inconvenience caused.This is our introduction to the oeuvre of Dr Li Jia. Now care is required when counting how many of her papers have been discussed at PubPeer (and the Researchgate archives are little better), for there are some quite separate names in Chinese which become 'Li Jia' when romanised by the Pinyin system. Here we are concerned with the Dalian Medical Hospital cancer researcher, who received her Ph.D in 2006 - the basis for her initial papers (seven of them with her Ph.D advisor as main author and herself as first author). That Li Jia.
Advised by Professor Jianing Zhang, a Kyoto University-trained Doctor of pharmaceutical sciences, Li Jia received her PhD on Pathology and Pathophysiology from Dalian Medical University in 2006, with her PhD thesis titled “Effects of CD147 glycosylation on the lymphatic metastasis in murine hepatocarcinoma cells lines”. It won a title of distinguished PhD thesis of the Liaoning Province in 2007. After her PhD, Li Jia remained in her university and became full professor in August 2008, her numerous research grants established her academic success and earned her an own lab and a big team.
What draws me to this oeuvre is the breadth of fake-data styles it encompasses. The versatility is impressive. Future conferences on Data Integrity could use it for an overview of the range of graphical techniques available for inventing results in cell biology. In fact the sheer abundance of material to include has forced me to be systematic, and to arrange everything along a time-line. I have limited its scope to the decade 2006-2015, with 27 papers receiving post-publication review at PubPeer (the two retractions, both from 2014, are [22] and [23]).
The time-line is not comprehensive; it features images that tie everything together by occurring more than once. There was no space for exuberant but one-off fabrications, such as Figure 3B from [3] in which hepatocarcinoma cells multiply to form colonies in nutrient agar (peer-reviewers were not concerned that identical colonies arose independently, such as the Mickey Mouse silhouette marked with a blue trapezoid in the first and third panels).
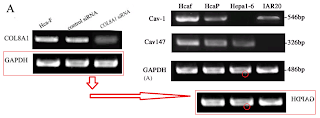
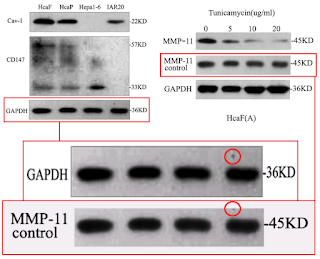
Over to the left of the chronology I have arranged the various manifestations of Western blots. The emphasis is on the metamorphoses of a small number of loading controls, the labels on the lanes changing according to the exigencies of different experiments. Now one school of thought regards loading controls as a needless, time-wasting formality, and relies on the process of harvesting cell contents to capture the same number of cells and yield the same concentration of proteins for each experimental situation (i.e. for each lane in the blot) - making it nugatory to control for that concentration. Jia was evidently of that persuasion.
However, that defense does not apply to the "swoosh" series of metamorphoses (from papers #3, #5 and #7), in which the recurring blot serves as empirical evidence for the three substantive proteins MMP-11, COL8A1 and VEGF-A.
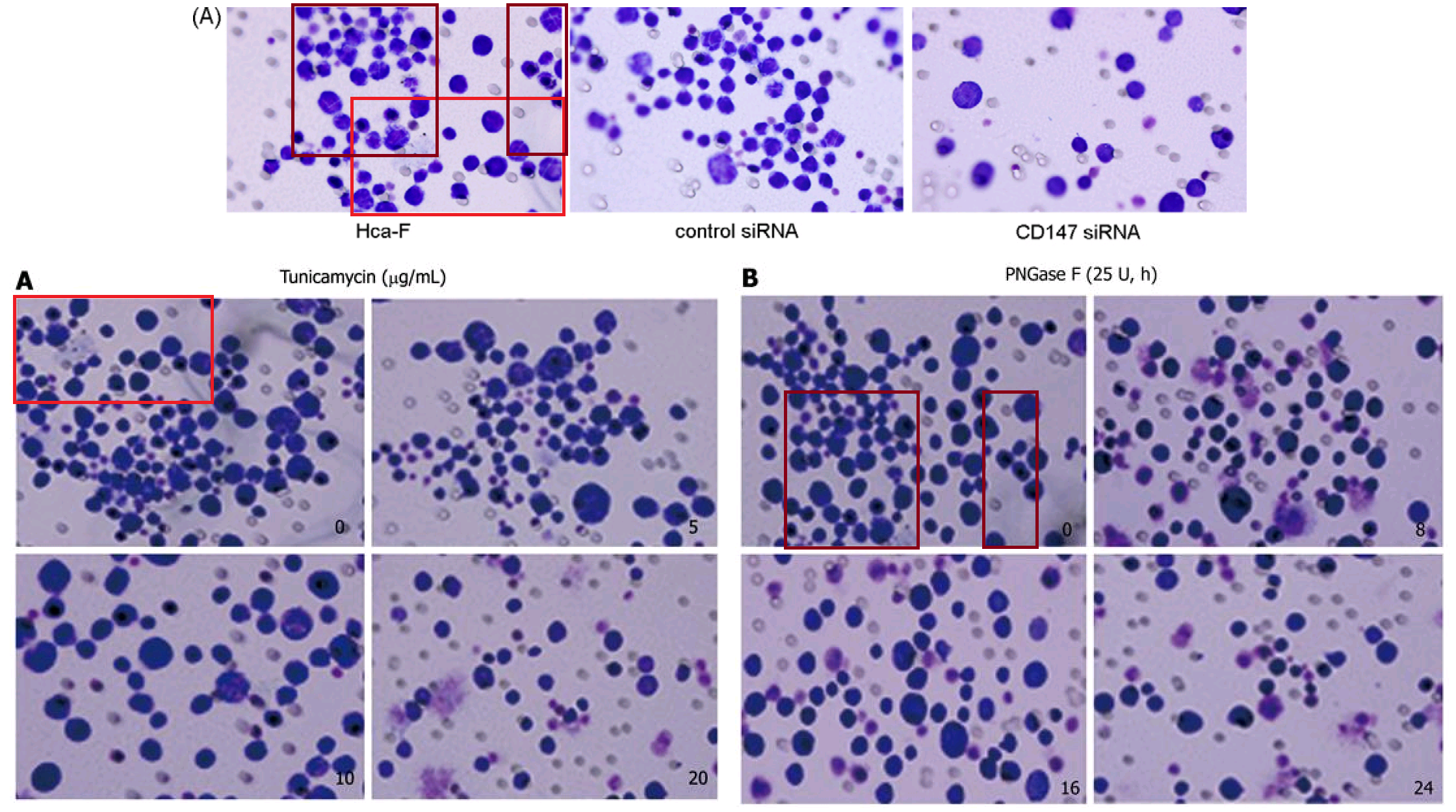
Apparently the same slice of gel can be a GAPDH control from different cell-lines in [1], or Matrix Metalloprotease-11 expression in the same cell-line exposed to different concentrations of tunicamycin, in [2]. None of this is promising for the Ph.D thesis source. Nor is it a good advertisement for supervisor and main author Jianing Zhang, who had left Dalian Medical University in 2013 and took the position of the Dean of the School of Life Science and Medicine in a neighbouring, but much more prestigious Dalian University of Technology.
That second manifestation as MMP-11 opens up further vistas of exploration when we look at its nominal loading control. It overlaps with other controls in the same paper, and they with others still... until we can reconstruct a single Ur-blot with nine lanes, from which various segments were extracted to provide the GAPDH controls for five separate blots, comparing multiple gamuts of conditions and spanning papers [1], [2] and [9].
No reconstruction is needed for a 12-lane GAPDH blot, which was shown in full in Figure 2E of [3], ensuring stability over a time-course.


Short excerpts from it can be found controlling for other trials in [3], [5] and [7] (where we met them already), and [11].
However, new material did enter the repertoire after 2010. One eight-lane band will always be "lipstick traces" to me. Various fragments featured as Figure 5A of [12], as 6A of [17], and four times in Figure 5 of [16].
The time-line includes a 2011 gap between [10] and [11], when Jia spent a year in Japan on a research fellowship (May 2010-May 2011) on the basis of her promising youthful productivity. Evidently this provided fresh blots, among them a second six-lane segment featured in overlapping segments in [11], [15], [16] and [17]... I like to think of it as the "charm bracelet".


In total, counting Figure 1D of [11] as well as 6B, the charms appeared five times. As its debut it took the form of the substantive protein B4GALT1 to justify a crucial claim of the paper: that the expression of that gene was blocked by a targeted shRNA. In that manifestation it acquired a GAPDH loading control of its own, which was less versatile, occurring only twice there and again in [12].


From Western Blots we shift to a genre grouped on the extreme right of the time-line. Here the molecules of interest - to be separated by molecular weight and labelled - are DNA segments, transcribed from RNA in the cellular extracts using Reverse Transcriptase, then amplified with PCR. Repurposed Northern Blots are not a novelty, and readers may recall them coming under scrutiny in previous blogging, but Jia took them to a new level. Prepare yourself for flotillas of kayaks and Canadian canoes parading across the screen, like a Mohawk war party in a Fenimore Cooper novel.
Two strands of repurposing can be discerned. The less active strand is distinguished by the crumpled prow (or stern) of one of the kayaks, and comprises only [3], [4], [6] and [13].


The "core blot" of the second strand is recognisable by the wee peg below the keel of one kayak and by the dot above another one (I surmise that to be the vanguard of the war party, and the leader has raised a small balloon, in the manner of a tour-group leader, to stop the others wandering off and becoming separated). It was cropped to two bands and three lanes for its debut in [1], and to a three-by-four form for [2]
This blot had already been manipulated when it first appeared outside Jia's thesis: the upturned kayak at the top right corner is a fabrication, spliced in. Anyway, these distinguishing marks allow us to trace the blot through its variants and permutations in [3], [4], [5], [6], [7] and [8]. Sometimes the kayaks are merely flipped vertically [5] or horizontally [7]:

Conversely, in [4] the manipulations reach delirious heights of ransom-note rearrangement to create eight lanes.


The two strands meet in Figure 1A of [6], which is a veritable stamp collection. Even a Helpful Diagram does not really make clear how much the core blots were sliced up and reconfigured.


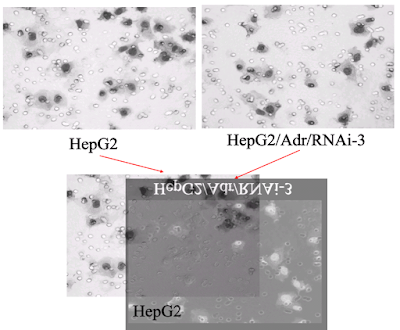
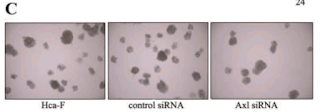
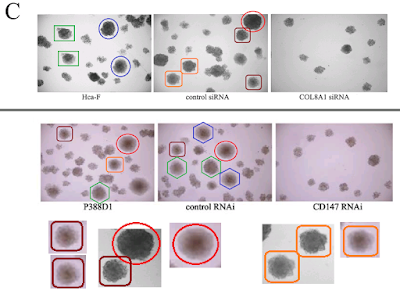
Let's go back to the Colony Growth assay, and to a group of images recycled across [4], [7], [8] and [10]. Everything can be improved with googly eyes, but also with duplicated colonies. Among other discoveries, it emerged that COLA81 manipulations affect the proliferation of Hca-F cells in exactly the same way as CD147 manipulations affect P388D1 cells... even producing identical colonies (in multiple copies). To my mind this phenomenon deserves a paper in itself.
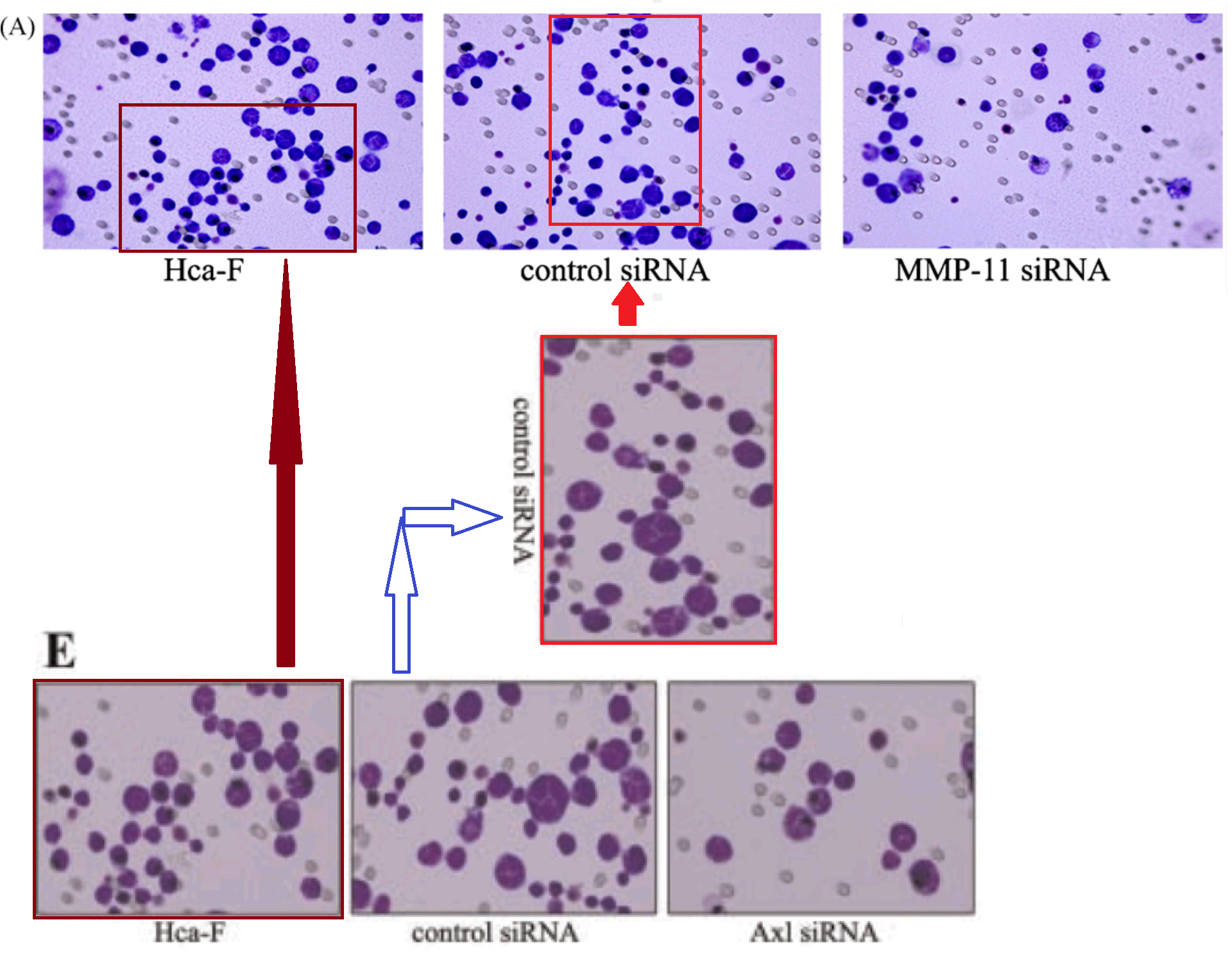
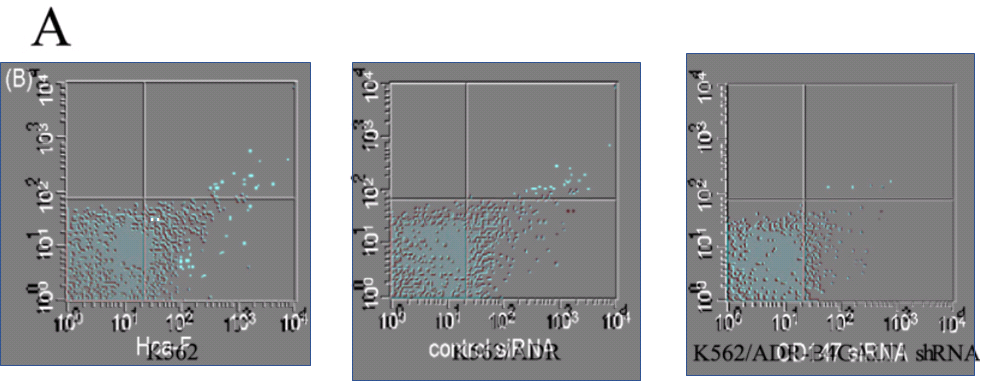
We have already encountered the Invasion / Migration Assay. There is some visual variety in its applications, depending on the specific lineage of cancer cells that had squeezed through pores in the filter. The software enhancement of the images is constant, though, and B. Mannanilyticus noted a number of cloned cells in Figure 3B of [16] (see also 4B).


Some of the repetitions stem from the fact that the upper-right and lower-right panels of 3B are essentially the same, as can be seen when one of them is colour-inverted and superimposed on the other, so that identical areas cancel out. The areas of failed cancellation are where cells were added to one panel, or the other, but not both.
We should also pay attention to the upper-left panel of 3B, for it reappeared in [19] as the top right of Figure 5(b), minus the Photoshop improvements (again, an inversion and superimposition clarify the areas of difference). The repeating sections are confined to other panels of 5(b), and to look at them closely is to be reminded of the crude faces in Appel's "Vragende Kinderen" paintings.


But we cannot linger on those images for I am impatient to move on to a group of invasion-assay results which may well be hommages to Yayoi Kusama and demanded their own thread in the time-line. Overlapping sections of panels reappear between a half-dozen papers, stretching across seven years, purportedly representing treatment with different siRNAs to discourage the cells' mobility. This series began in 2007 with Figure 5A of [3], where the most obvious overlaps are with 2E from [10].
Note that a prominent cell was inserted for the [10] version (or perhaps erased for the [3] version), presumably for aesthetic reasons. The authors continued to improve the images with more dots and manipulations, as if in the thrall of a gaes or whatever is the opposite of trypophobia, and the series build to a crescendo in [13] (2012)...
... then reached a dizzying apotheosis of artificiality in [18] (2013).




The failure of the PLoS-One reviewers to recognise the creativity poured into these figures must have been galling. Anyway, the links of re-use / alteration between papers are spelled out in the PubPeer threads, and rather than take up more space, I have prepared a little diagram to help interested readers follow the routes from one to another.
We are nearly finished with Invasion / Migration Assay images. The ones featured in [24], [25] and [26] are close to the angular cranes-in-flight profiles with which we began, though because they are purple I'll compare them to tropical fish in aquarium tanks. I lied when I wrote that "the software enhancement of the images is constant", for there is nothing amiss with these examples... except their re-use within and between papers, differently labelled.
One visual motif remains to tie these papers together. The FACS-plot strand stretches practically the length of this sample, from [3] to [27], though only five papers take part.
Because the three panels of 3B from [5] portray the protein-expression distributions of murine hepatocarcinoma cells, while panels 1, 2 and 4 of Figure 3A from [7] involve human leukemia cells - measured under different conditions - we expect them not to cancel out when panel is superimposed upon panel. But prepare for disappointment:
The repetitions are not complete; some points have been added or scraped away (each point indicating a single cell), as if an experimental data-file had been plotted with a different choice of filtering parameters, or simply revisited in Photoshop. As for the third panel of 3A, it is an unnatural chimera, formed by grafting two half-panels together.
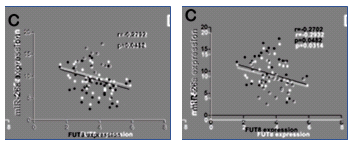
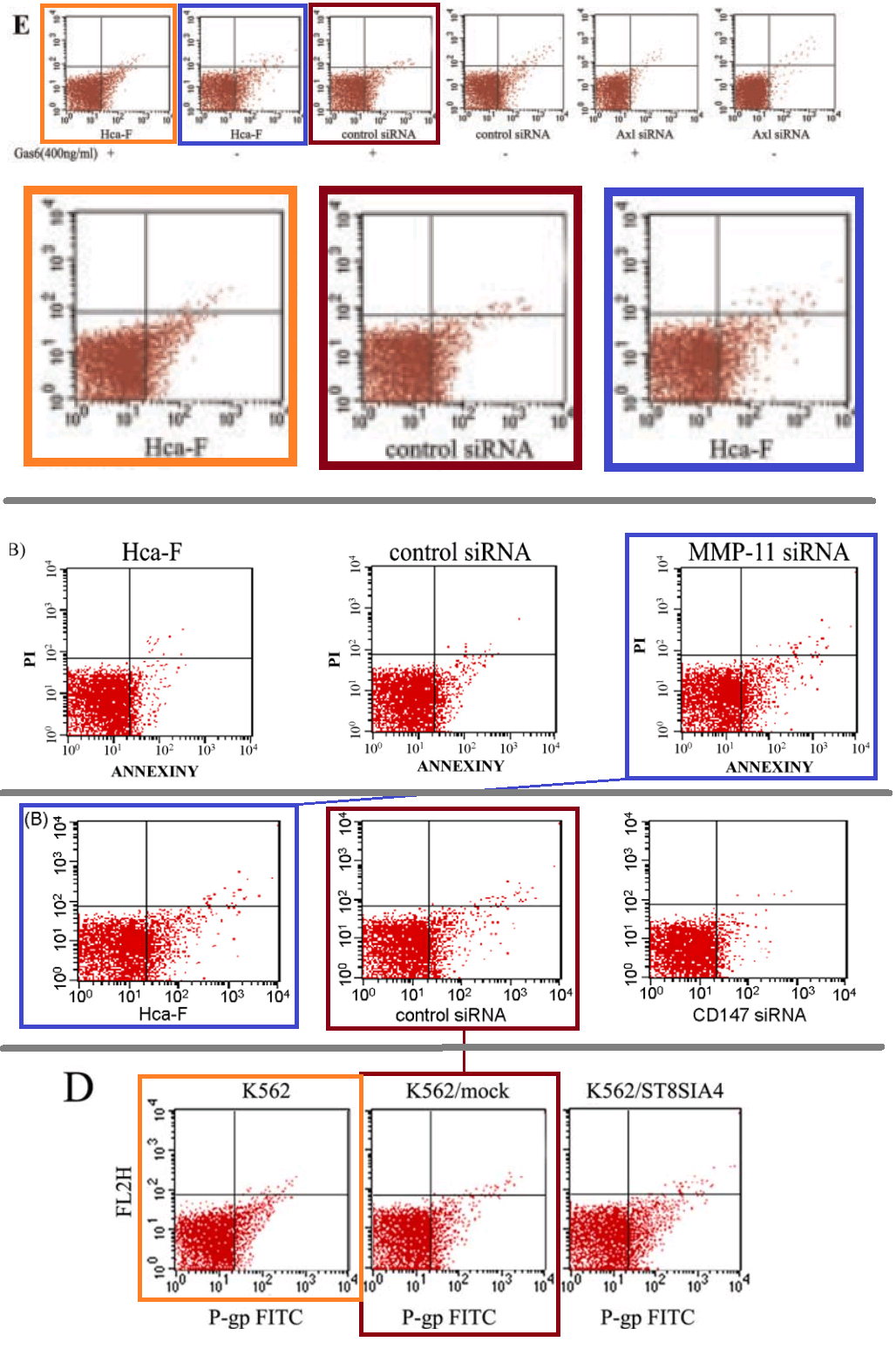 A single color-coded diagram captures the almost-repetitions across Figures 4E from [10], 4B from [3], 3B from [5] and 6D from [27].
A single color-coded diagram captures the almost-repetitions across Figures 4E from [10], 4B from [3], 3B from [5] and 6D from [27].After 2015, Li Jia's research shifted direction towards the area of micro-RNA and its role in intra- and inter-cellular regulation. [28] is "Comprehensive N-glycan profiles of hepatocellular carcinoma reveal association of fucosylation with tumor progression and regulation of FUT8 by microRNAs", from 2016. [29] is "Long non-coding RNA HOTAIR promotes osteoarthritis progression via miR-17-5p/FUT2/β-catenin axis".
A feature of this newly-popular miRNA genre is its strong visual conventions. Papers follow a template, and as well as Western Blots and FACS plots, readers expect to see immunofluorescent staining and scatterplots of miRNA levels against cancer biomarkers.
Fig 4C of [28]: Not entirely clear on the "independent measurements" aspect of scatterplots
As part of that shift, her laboratory left behind the egregious displays of forged results. Another 10 threads at PubPeer examine her output since 2015, but with some exceptions they concern trivial image duplications that can be corrected with an embarrassed Erratum notice and a Does-not-affect-the-Conclusions. Indeed, four of the affected papers have been recently amended (coinciding with the Retraction with which we began). Note, too, that these 10 papers are only a fraction of the group's productivity, with 11 papers published in 2017 and nine in 2018.
"With some exceptions". Figure 5C from [29].
It may be that Li Jia does not need to make up results now that she has students of her own. Once you make it you no longer have to fake it. It may also be that I spent too much time on over-thinking this. Arguably there is nothing exceptional about this body of work, though it does conveniently bring together a diverse range of ways in which data manipulation can manifest.
Here are some out-takes. The task of finding the source(s) is left as an exercise for the reader.

[1] "Deglycosylation of CD147 down-regulates Matrix Metalloproteinase-11 expression and the adhesive capability of murine hepatocarcinoma cell HcaF in vitro", IUBMB Life (2006). Li Jia, Huimin Zhou, Shujing Wang, Jun Cao, Wei Wei, Jianing Zhang. (PubPeer here)
[2] "Caveolin-1 up-regulates CD147 glycosylation and the invasive capability of murine hepatocarcinoma cell lines", International Journal of Biochemistry & Cell Biology (2006). Li Jia, Shujing Wang, Huimin Zhou, Jun Cao, Yichuan Hu, Jianing Zhang. (PubPeer here)
[3] "siRNA targeted against matrix metalloproteinase 11 inhibits the metastatic capability of murine hepatocarcinoma cell Hca-F to lymph nodes", International Journal of Biochemistry & Cell Biology (2007). Li Jia, Shujing Wang, Jun Cao, Huimin Zhou, Wei Wei, Jianing Zhang. (PubPeer here)
[4] "CD147 regulates vascular endothelial growth factor-A expression, tumorigenicity, and chemosensitivity to curcumin in hepatocellular carcinoma", IUBMB Life (2007). Li Jia, Huaxin Wang, Shuxian Qu, Xiaoyan Miao, Jianing Zhang. (PubPeer here)
[5] "CD147 depletion down-regulates matrix metalloproteinase-11, vascular endothelial growth factor-A expression and the lymphatic metastasis potential of murine hepatocarcinoma Hca-F cells", International Journal of Biochemistry & Cell Biology (2007). Li Jia, Jun Cao, Wei Wei, Shujing Wang, Yunfei Zuo, Jianing Zhang.. (PubPeer here)
[6] "Expression of CD147 mediates tumor cells invasion and multidrug resistance in hepatocellular carcinoma", Cancer Investigation (2008). Li Jia, Henggui Xu, Yongfu Zhao, Lili Jiang, Jingda Yu, Jianing Zhang. (PubPeer here)
[7] "siRNA-targeted COL8A1 inhibits proliferation, reduces invasion and enhances sensitivity to D-limonence treatment in hepatocarcinoma cells", IUBMB Life (2009). Yongfu Zhao, Li Jia, Xiaoyan Mao, Henggui Xu, Bo Wang, Yongji Liu. (PubPeer here)
[8] "Silencing CD147 inhibits tumor progression and increases chemosensitivity in murine lymphoid neoplasm P388D1 cells", Annals of hematology (2009). Li Jia, Wei Wei, Jun Cao, Henggui Xu, Xiaoyan Miao, Jianing Zhang. (PubPeer here)
[9] "Knockdown of Caveolin-1 by siRNA Inhibits the Transformation of Mouse Hepatoma H22 Cells In Vitro and In Vivo", Oligonucleotides (2009). Shujing Wang, Li Jia, Huimin Zhou, Wei Shi, Jianing Zhang. (PubPeer here)
[10] "Differential expression of Axl in hepatocellular carcinoma and correlation with tumor lymphatic metastasis", Molecular Carcinogenesis (2010). Ling He, Jianing Zhang, Lili Jiang, Changgong Jin, Yongfu Zhao, Guang Yang, Li Jia. (PubPeer here)
[11] "Differential expression of Axl and correlation with invasion and multidrug resistance in cancer cells", Cancer Investigation (2012). Yongfu Zhao, Xiance Sun, Lili Jiang, Fengjuan Yang, Zhaohai Zhang, Li Jia. (PubPeer here)
[12] "Glycomic alterations are associated with multidrug resistance in human leukemia", International Journal of Biochemistry & Cell Biology (2012). Zhaohai Zhang, Yongfu Zhao, Lili Jiang, Xiaoyan Miao, Huimin Zhou, Li Jia. (PubPeer here)
[13] "Effect of enhanced expression of COL8A1 on lymphatic metastasis of hepatocellular carcinoma in mice", Experimental & Therapeutic Medicine (2012). Zhen-Hai Ma, Jin-Hui Ma, Li Jia, Yong-Fu Zhao . (PubPeer here)
[14] "Axl glycosylation mediates tumor cell proliferation, invasion and lymphatic metastasis in murine hepatocellular carcinoma", World Journal of Gastroenterology (2012). Ji Li, Li Jia, Zhen-Hai Ma, Qiu-Hong Ma, Xiao-Hong Yang, Yong-Fu Zhao". (PubPeer here)
[15] "B4GALT1 gene knockdown inhibits the hedgehog pathway and reverses multidrug resistance in the human leukemia K562/adriamycin-resistant cell line", IUBMB Life (2012). Huimin Zhou, Zhaohai Zhang, Chunqing Liu, Changgong Jin, Jianing Zhang, Xiaoyan Miao, Li Jia. (PubPeer here)
[16] "Glycogenes mediate the invasive properties and chemosensitivity of human hepatocarcinoma cells", International Journal of Biochemistry & Cell Biology (2013). Rui Guo, Lei Cheng, Yongfu Zhao, Jianing Zhang, Chunqing Liu, Huimin Zhou, Li Jia. (PubPeer here)
[17] "Functional roles of glycogene and N-glycan in multidrug resistance of human breast cancer cells", IUBMB Life (2013). Hongye Ma, Xiaoyan Miao, Qiuhong Ma, Wenzhi Zheng, Huimin Zhou, Li Jia. (PubPeer here)
[18] "Modification of glycosylation mediates the invasive properties of murine hepatocarcinoma cell lines to lymph nodes", PLoS ONE (2013). Zhaohai Zhang, Jie Sun, Lihong Hao, Chunqing Liu, Hongye Ma, Li Jia. (PubPeer here)
[19] "Axl gene knockdown inhibits the metastasis properties of hepatocellular carcinoma via PI3K/Akt-PAK1 signal pathway", Tumor Biology (2014). Jingchao Xu, Li Jia, Hongye Ma, Yanping Li, Zhenhai Ma, Yongfu Zhao. (PubPeer here)
[20] "Reversal effect of ST6GAL 1 on multidrug resistance in human leukemia by regulating the PI3K/Akt pathway and the expression of P-gp and MRP1", PLoS ONE (2014). Hongye Ma, Lei Cheng, Keji Hao, Yanping Li, Xiaobo Song, Huimin Zhou, Li Jia. (PubPeer here)
[21] "ST6GalNAcII mediates the invasive properties of breast carcinoma through PI3K/Akt/NF-κB signaling pathway", IUBMB Life (2014). Dongliang Ren, Li Jia, Yanyan Li, Yanxin Gong, Chen Liu, Xu Zhang, Ning Wang, Yongfu Zhao. (PubPeer here)
[22] "Effect of ST3GAL 4 and FUT 7 on sialyl Lewis X synthesis and multidrug resistance in human acute myeloid leukemia", Biochimica et Biophysica Acta (2014). Hongye Ma, Huimin Zhou, Peng Li, Xiaobo Song, Xiaoyan Miao, Yanping Li, Li Jia. (PubPeer here)
[23] "Modification of sialylation mediates the invasive properties and chemosensitivity of human hepatocellular carcinoma", Molecular & Cellular Proteomics (2014). Yongfu Zhao, Yanping Li, Hongye Ma, Weijie Dong, Huimin Zhou, Xiaobo Song, Jianing Zhang, Li Jia.(PubPeer here)
[24] "Phyllodes tumor of the breast: role of Axl and ST6GalNAcII in the development of mammary phyllodes tumors", Tumor Biology (2014). Dongliang Ren, Yanyan Li, Yanxin Gong, Jingchao Xu, Xiaolong Miao, Xiangnan Li, Chen Liu, Li Jia, Yongfu Zhao. (PubPeer here)
[25] "Axl mediates tumor invasion and chemosensitivity through PI3K/Akt signaling pathway and is transcriptionally regulated by slug in breast carcinoma", IUBMB Life (2014). Yanyan Li, Li Jia, Dongliang Ren, Chen Liu, Yanxin Gong, Ning Wang, Xu Zhang, Yongfu Zhao. (PubPeer here)
[26] "Axl as a downstream effector of TGF-β1 via PI3K/Akt-PAK1 signaling pathway promotes tumor invasion and chemoresistance in breast carcinoma", Tumor Biology (2014). Yanyan Li, Li Jia, Chen Liu, Yanxin Gong, Dongliang Ren, Ning Wang, Xu Zhang, Yongfu Zhao. (PubPeer here)
[27] "α-2,8-Sialyltransferase Is Involved in the Development of Multidrug Resistance via PI3K/Akt Pathway in Human Chronic Myeloid Leukemia", IUBMB Life (2015). Xu Zhang, Weijie Dong, Huimin Zhou, Hongshuai Li, Ning Wang, Xiaoyan Miao, Li Jia. (PubPeer here)
[28] "Comprehensive N-glycan profiles of hepatocellular carcinoma reveal association of fucosylation with tumor progression and regulation of FUT8 by microRNAs", Oncotarget (2016). doi: 10.18632/oncotarget.11284. Lei Cheng, Shuhang Gao, Xiaobo Song, Weijie Dong, Huimin Zhou, Lifen Zhao, Li Jia (PubPeer here)
[29] "Long non-coding RNA HOTAIR promotes osteoarthritis progression via miR-17-5p/FUT2/β-catenin axis", Cell Death & Disease (2018). doi: 10.1038/s41419-018-0746-z Jialei Hu, Zi Wang, Yujia Shan, Yue Pan, Jia Ma, Li Jia (PubPeer here)





















No comments:
Post a Comment